Metabolomics
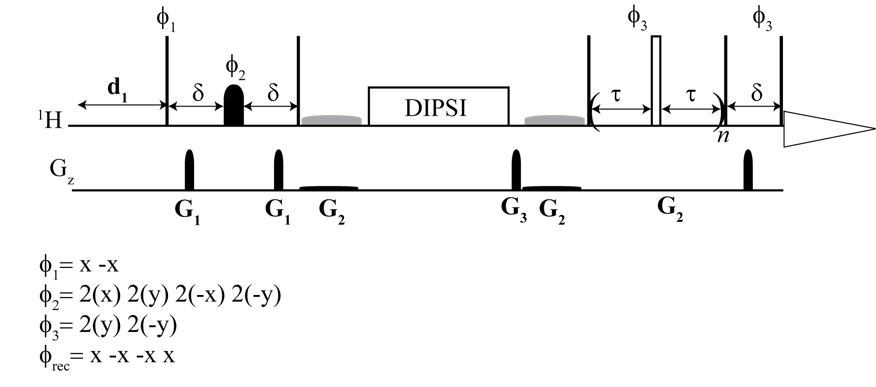
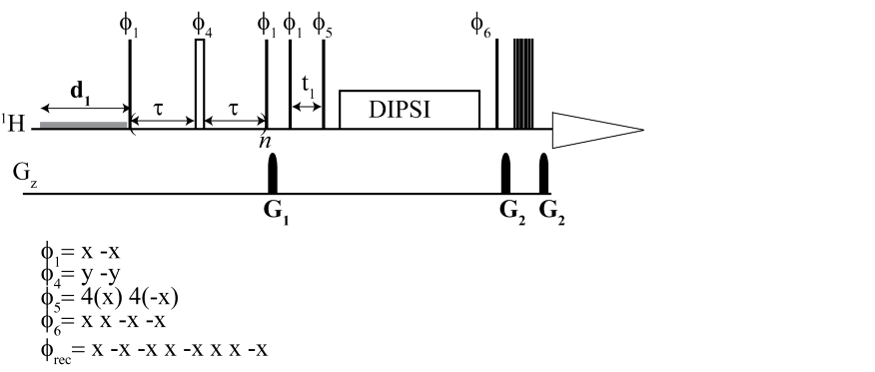
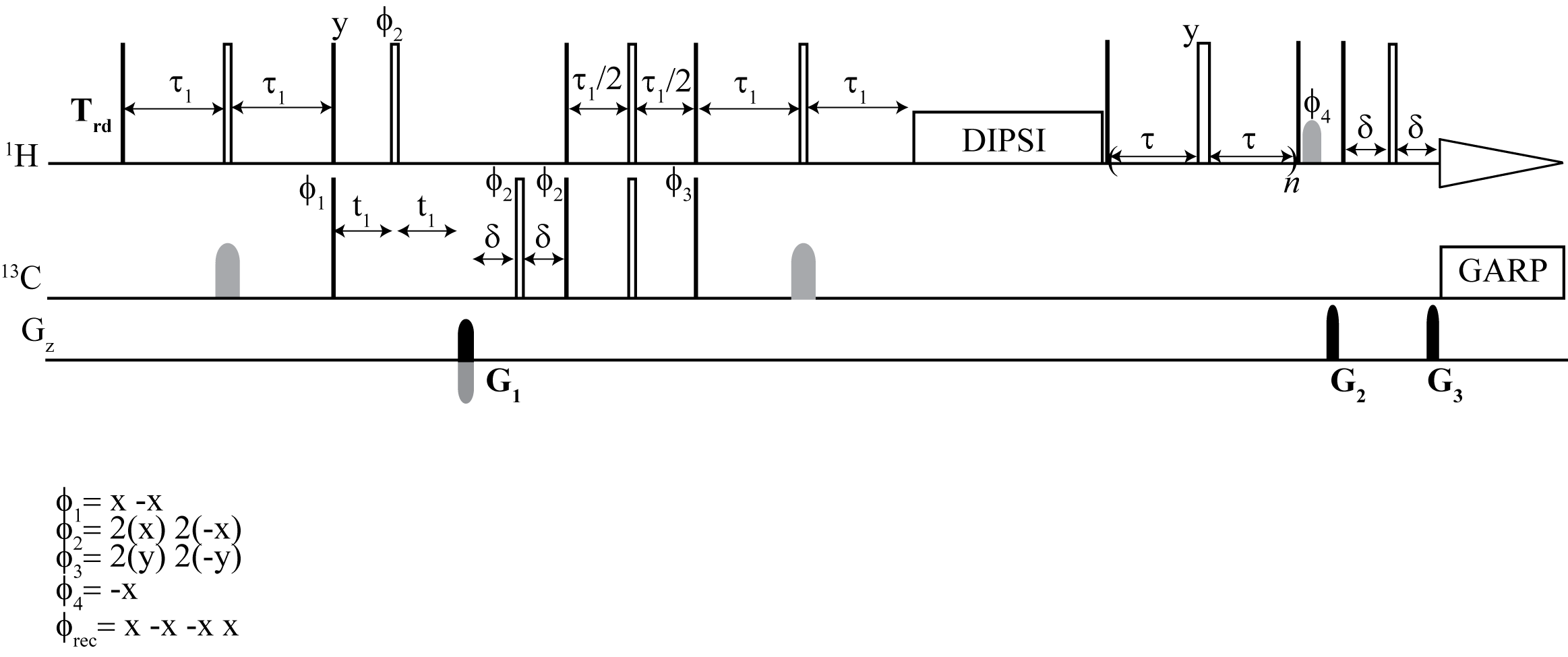
From the top: 1H CPMG TOCSY, 2D 1H–1H CPMG TOCSY, 2D 13C–1H CPMG HSQC TOCSY
Here, we implemented and validated a suite of selective and non-selective CPMG-filtered 1D and 2D TOCSY/HSQC experiments for metabolomics research. They facilitated the unambiguous identification of metabolites embedded in broad lipid and protein signals. The 2D spectra improved non-targeted analysis by removing the background broad signals of macromolecules.
We provide a complete set of experiments, which are CPMG and TOCSY based for both targeted and non-targeted analysis. The main aim was unambiguous identification and metabolic profiling. We combined the advantage of selective excitation for targeted analysis, multidimensional resolution for both proton and carbon, along with CPMG and TOCSY. We implemented TOCSY on HSQC, the most sensitive heteronuclear 2D experiment for metabolomics, along with CPMG to retain the benefit of enhanced carbon resolution. The experimental data obtained using this technique leave all opportunities open for advanced pattern matching algorithms for assignment.
Abstract
Here, we implemented and validated a suite of selective and non-selective CPMG-filtered 1D and 2D TOCSY/HSQC experiments for metabolomics research. They facilitated the unambiguous identification of metabolites embedded in broad lipid and protein signals. The 2D spectra improved non-targeted analysis by removing the background broad signals of macromolecules.